| |||||||||||||
|
|
Journals 2006/2007Tamara Browning
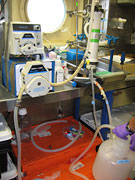
August 29, 2006 When I first met Karla Heidelberg prior to embarking for the second leg of the cruise, she was loading her equipment onto the ship. I was intrigued by the huge plastic bottles that she had brought along with her. There were four 50 liter screw top bottles and a large plastic barrel about the size of a garbage can that held 200 liters. She also had quantities of plastic tubing, two specialized pumps and a three-legged filtering device with eight handles that had to be tightened using a wrench. Judging by the equipment her research was very different from the bongo based sampling that the rest of us were involved with. It was not until a few days later when Karla explained to me what she was doing, that I grasped how "cutting edge" her area of research was. The large bottles had misled me since it turned out that her interests lay at the other end of the size scale, with the microbes - bacteria and viruses - that inhabit the surface of the ocean. She was onboard to continue with an investigation that involved sampling water from several sites in the Gulf of Maine in order to learn more about the community of microorganisms that inhabit this part of the ocean. Extracting the microorganisms from the water was a time consuming process that lasted about 20 hours. It involved pumping a 200 liter sample of sea surface water through a set of three filters with progressively smaller openings. Bacteria would be trapped on the third filter with the smallest mesh size, but viruses are so tiny that they would just flow right on through. So the final stage of the collection process involved passing the virus laden water through an instrument called a "50Kda cut-off tangential flow filter" that basically removed most of the water leaving behind the viruses concentrated together in about half a liter of water.
Back at the lab, DNA will be extracted from the microorganisms and subjected to a novel technique called shotgun genome sequencing". Ultimately this will provide information about what types of genes are present and active in the community of bacteria and viruses that live in the Gulf of Maine. This information will be added to the genetic data bank that Karla and others are currently developing using microbial communities obtained from ocean sites around the world. Until very recently these microscopic inhabitants of the ocean have escaped the attention of scientists, since most of them were impossible to culture in the lab. Now using these new techniques researchers were beginning to unravel the genetic potential locked up in the microbes that inhabit the ocean surface. |
||||||||||||